Putting it all together
Overview
Teaching: 45 min
Exercises: 50 minQuestions
How do RMarkdown, Git, and Open Science Framework work together to produce a reproducible data analysis?
Objectives
To produce a simple example of a reproducible workflow
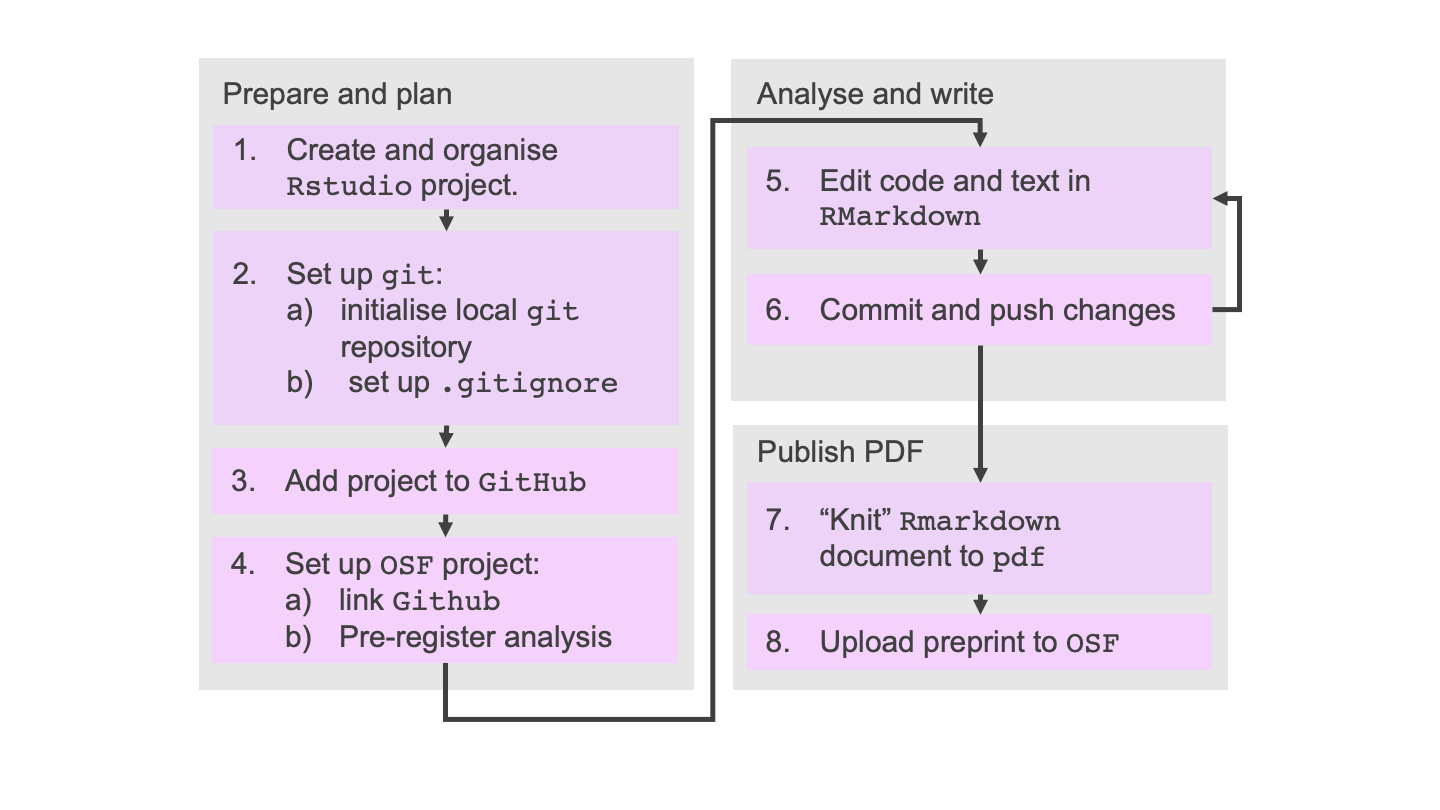
Putting it together
We’ll now be putting everything that we’ve leared together and practicing the workflow in its entirety. As an example project we’ll be building on the analysis of the Old Faithful data which was started in episode 3.
Prepare and plan
Step 1: Create and organise RStudio project
Create a brand new project (in RStudio cloud or RStudio local) using the instructions
episode 2. Call the project old_faithful.
Place the project somewhere convenient for you e.g., Desktop/old_faithful.
Steps 2 and 3: Set up Git and Github
Initialize a local git repository (in RStudio cloud or RStudio local) using the instructions
episode 4. You should call
the repository old_faithful.
Copy the old_faithful_updated.Rmd analysis from episode 3.
Rename old_faithful_updated.Rmd -> old_faithful_eda.Rmd. This is to better reflect what the
document is about (EDA = exploratory data analysis) and to differentiate it from other files that we will create.
If you’re using RStudio cloud you can’t copy files between project. Instead upload the solutions version available here.
Make sure you’re ‘ignoring’ pdfs in your .gitignore file.
Commit and push this.
Step 4: Preregister the analysis on the Open Science Framework.
We’re now going to write down our analysis plan and preregister a simple hypothesis. This will be done in three steps:
- Create an Open Science Framework (OSF) project.
- Write an analysis plan (a plan will be provided).
- Upload documents and preregister the project.
1. Create an OSF project
- Sign up, if you haven’t already, for an osf account.
- On the dashboard click
Create new project. Call it ‘Old Faithful’ and thenGo to project.
2. Create hypothesis and analysis plan
Our hypothesis is:
There are 2 distinct types of eruption from Old Faithful: 1) short frequent eruptions, 2) long infrequent eruptions.
We shall test it by clustering the data using a Gaussian mixture model (more information here) with 1 and 2 components. The true number of components will be given by the model with the smallest Integrated Complete Data Likelihood (ICL) criterion. The model will be fit in the R package RMixmod.
(Don’t worry if you don’t understand this - all the materials will be provided!)
- Download the analysis plan here
- Commit and push changes.
Knitbothanalysis_plan.Rmdandold_faithful_eda.Rmdfiles to pdfs.- Upload both
analysis_plan.pdfandold_faithful_eda.pdfdocuments to the OSF project. To do this navigate toFiles, click onOSF Storage (United States). TheUploadbutton should appear and you can use this to upload files.
3. Preregister plan and project.
The OSF instructions for registering projects can be found here. Specific instructions for this project are given here:
- Click
Registrations>New registration - Select
Open-Ended Registrationand then selectCreate draft. - Fill in the required
Metadata:.- Description: An investigation into the Old Faithful geyser
- License:
CC-By Attribution 4.0 International. - Subjects:
Physical Sciences and Mathematics.
- Click
Nextthen fill in theSummary:Summary: The attached file analysis_plan.pdf contains our project description, hypothesis and analysis plan.- Select the
analysis_plan.pdfdocument.
- Now click
Reveiw. Review your submission and when you’re satisfied clickRegister. - You can now click
Make registration public immediatelythenSubmit.
The registration is now pending approval by the administrators of the project (i.e., you). If you navigate back to the Registrations tab of the project and click on the link next to Pending registration, it will bring you to your registration page. This contains the information you entered when registering the project, as well as a snapshot of the project contents, i.e., analysis_plan.pdf and old_faithful_eda.pdf.
You will get an email asking you to approve or cancel this preregistration. You should cancel this preregistration after the course.
Analysis and write
Steps 5 and 6: Perform analysis and commit changes
- Download the report template from here.
- You will need to install
Rmixmod. You will be askedDo you want to install from sources the package which needs compilation? (Yes/no/cancel)- chooseno. - Follow the instructions in the report template. You should make a commit and push after every couple of completed tasks in the report template. i.e. you should commit and push multiple times. This is good practice.
A completed report is available here should you need help.
Publish PDF
Step 7 and 8:
Knitthe completedreport_template.Rmdto a pdf.- Upload the pdf to the OSF project.
You have now successfully completed a fully reproducible piece of preregistered data analysis!
Both the preregistration and the final analysis report are all publicly available on the OSF.
Your code is available for anyone to use from your Github profile.
Conclusions
That concludes the course. In this last episode we have brought together all the elements learned in the previous episodes and applied it to an example analysis of the Old Faithful dataset. We used RStudio to create an project ( episode 2 ) to contain our Old Faithful analysis. Then we performed some exploratory data analysis and wrote an analysis plan using RMarkdown ( episode 3). From episode 1 we learnt about preregistration and so we preregistered our hypothesis with the OSF. We performed our intended analysis, backing up along the way using version control ( episode 4). We then published our report, along with our pre-registered analysis plan, on the OSF. In addition, because we used version control, our code was already published and available for use by others on Github.
Key Points
Open Science Framework (OSF) is a tool for organising and preregistering projects and sharing them on the web
Rstudio, Rmarkdown, Git and Github can be combined to create reproducible data analysis workflow